Liu, D., Yu, L., Wei, L., Yu, P., Wang, J., Hu, Z., Zhang, Y., Zhang, S., Yang, Z., Chen, G., Yao, X., Yang, Y., Zhou, Y., Wang, X., Lu, S., Dai, C., Yang, Q.Y. and Guo, L. (2021) BnTIR:an online transcriptome platform for exploring RNA‐seq libraries for oil crop Brassica napus. Plant biotechnology journal. https://doi.org/10.1111/pbi.13665
BnTIR used three different processes to integrate the Expression quantity data of 273 samples from 8 tissues ( root, stem, leaf, bud, flower, silique, silique_wall and seed ) on ZS11 in rapeseed cultivation , and these data were presented through eFP and Expression modules. The Expression module provides different retrieval modes , which can efficiently retrieve the expressions of a single gene in a single tissue , a single gene in multiple tissues , multiple genes in multiple tissues , and multiple genes in a single tissue , and provide abundant interactive result diagrams. Other bioinformatics tools have been developed,such as the JBrower browser, sequence fetch ,blast , transcription factor family query, and co-expression. BnTIR provides rapeseed researchers with a comprehensive gene expression profile and user-friendly visual interface, laying the foundation for understanding gene function at transcriptome level.
This is the navigation bar at the top of the BnTIR database, which contains eFP module、expression module、coxpression module、JBrower module、Tool module(blast、gene index、seq fetch、tf search)、Download module.
BnTIR provides eFP browser , allowing users to comprehensively view gene expression levels among 8 tissues at different stages of development.By entering "Data Source","Mode","Primary Gene ID","Secondary Gene ID","Signal Threshold" and clicking "Go",you will see the specific expression of genes in various tissues under controlled conditions.
On the Gene Expression search page, You have three input modes to choose from. 1)you can enter the full arabidopsis gene name or the rapeseed gene ID or the Arabidopsis gene ID. 2)you can enter a formatted region.(A01:150000..250000). 3)You can put the Bna gene ID in the other genome. and you will see the results of relevant genes.The results included basic information about the gene , its expression levels in different tissues, and some visual plot (phylotree、heatmap、linechart、boxplot plot) drawn from these expressions.
On co-expression page,by searching the gene number and selecting the depth and correlation coefficient (PCC), the co-expression network and basic gene information table of several genes can be obtained.
On JBrower page,the differences in gene expression levels in different tissues in different regions were visualized.
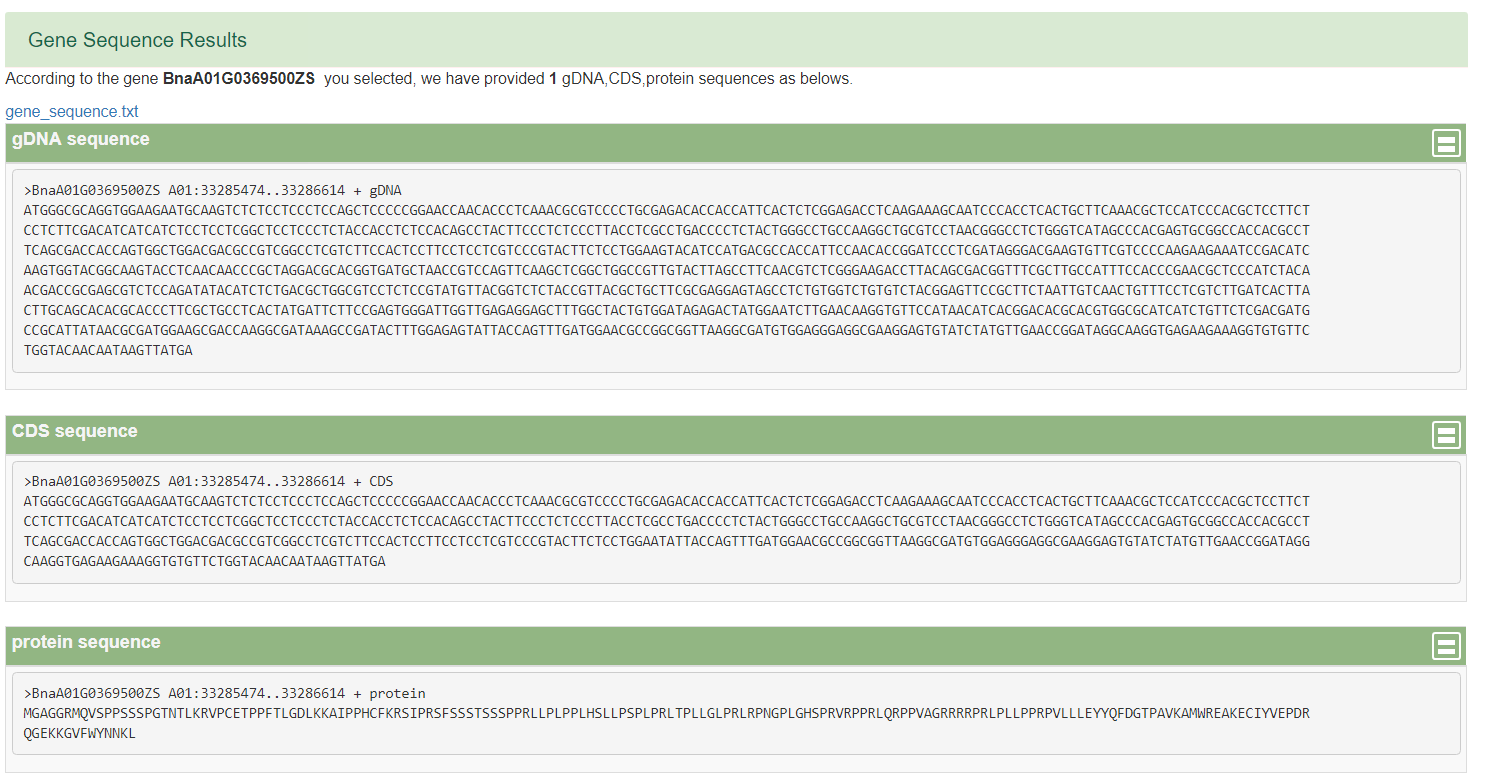
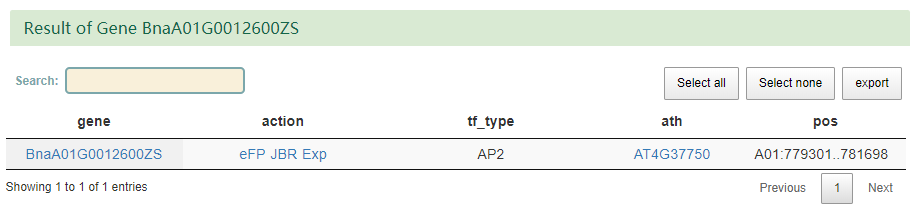
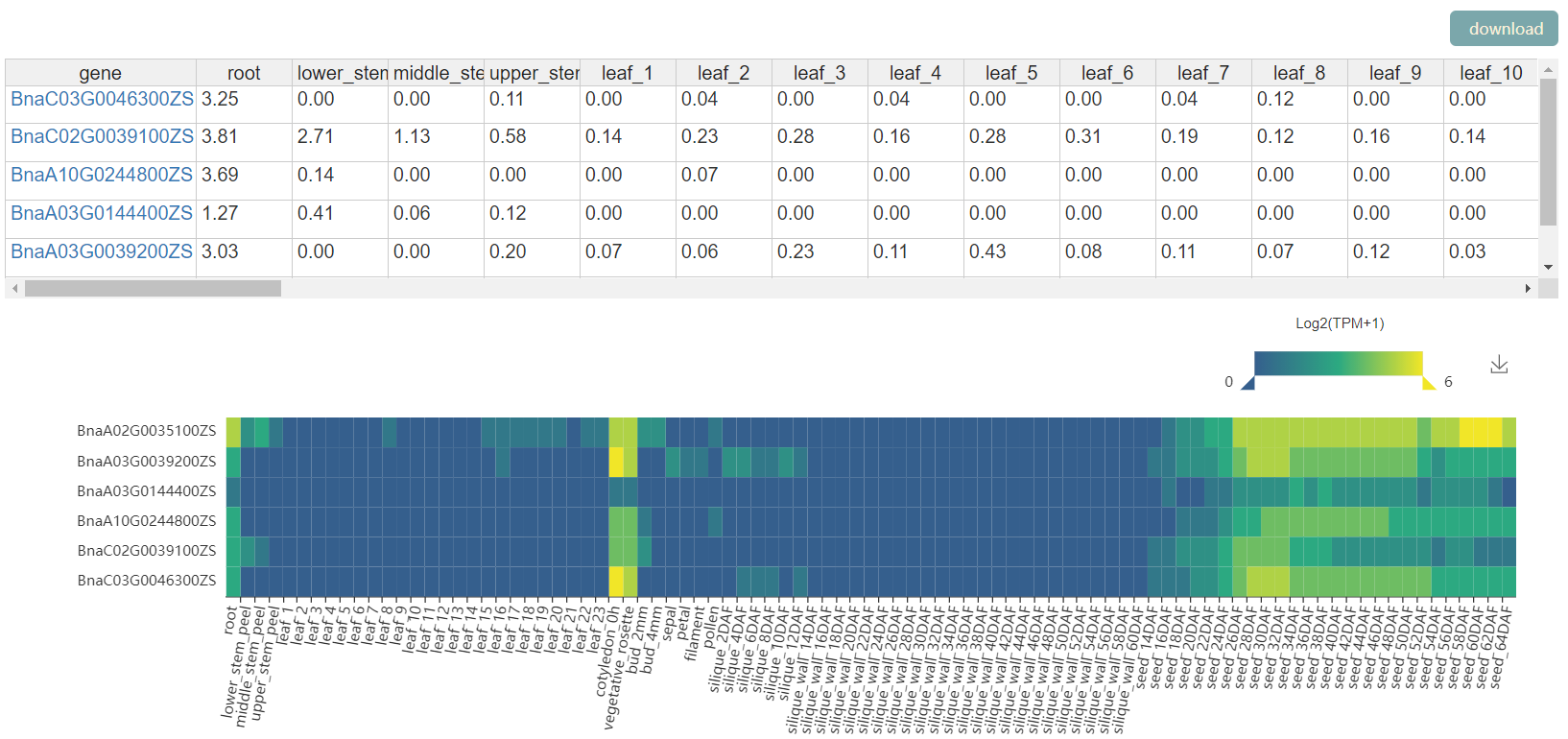
1)BnTIR provides Gene Index function.You can enter one or more Bna genes in a text box , or upload a file and submit it, and you'll see the id of the Bna gene ID in the rest of the genome. 2)BnTIR provides seq fetch function. Users can input rapeseed gene or a segment to obtain some basic information of the gene and corresponding gDNA,CDS and protein sequences, which can be copied or downloaded. 3)BnTIR provides TF function.Input mode one is to click the transcription factor family name to obtain all the gene information of the family, but to input the rape gene number (nine reference genomes can be used) or the name to obtain basic information and the family to which it belongs. 4)BnTIR provides heat mapping function.Users can enter any interested genes and tissues.At the same time, add some colors and calculate parameters. 5)BnTIR provides blast function.
2)
3)
4)
5)
On the Download page, you can enter a section or genes of interest, and then provided with a table of basic information and expression volume data.
On the methods page, you can see 91 sample indormation datatable、diagrams and basic descriptions of three sets of processes.
On the Contact page, we have provided the contact information as folows. If you have any questions or ideas, please feel free to contact us. email:yqy@mail.hzau.edu.cn Tel:027-87280877