Welcome to AMIR!
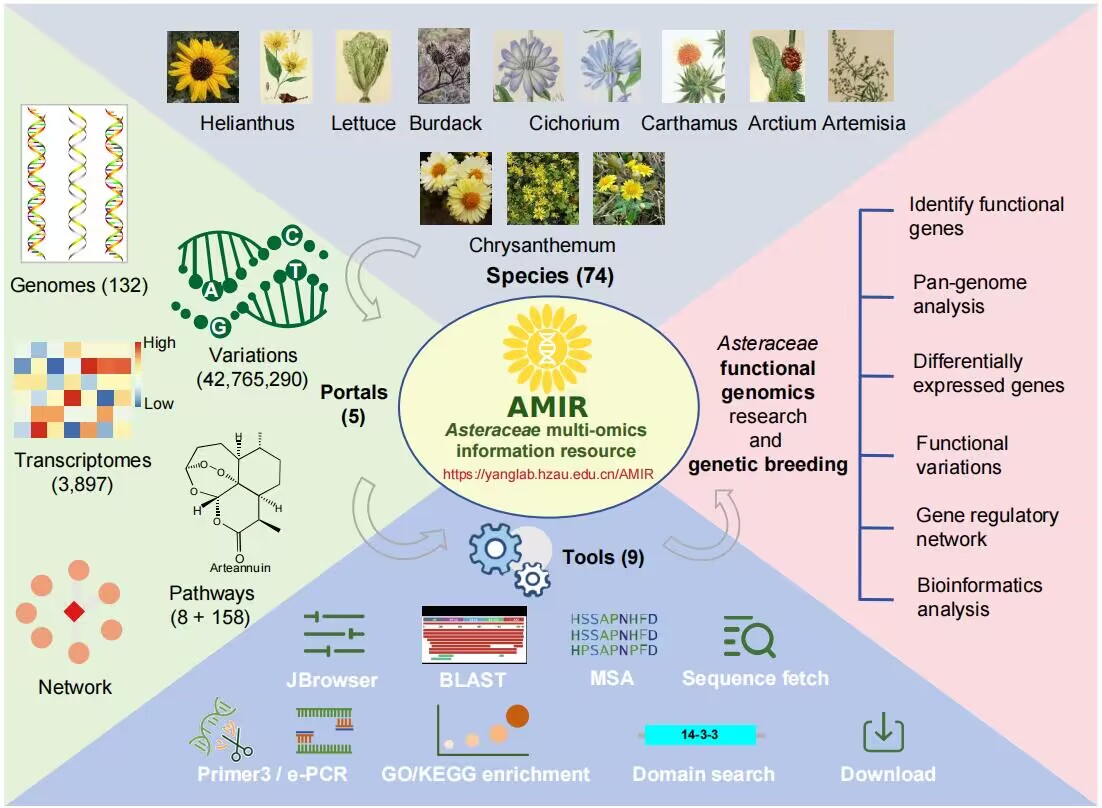
AMIR (Asteraceae Multi-omics Information Resource) is a well-organized, convenient graphical representation resource dedicated to collect, organize, display, and compare omics data in Asteraceae species.
- Retrieve 11,475,543 annotation data for individual genes or gene sets.
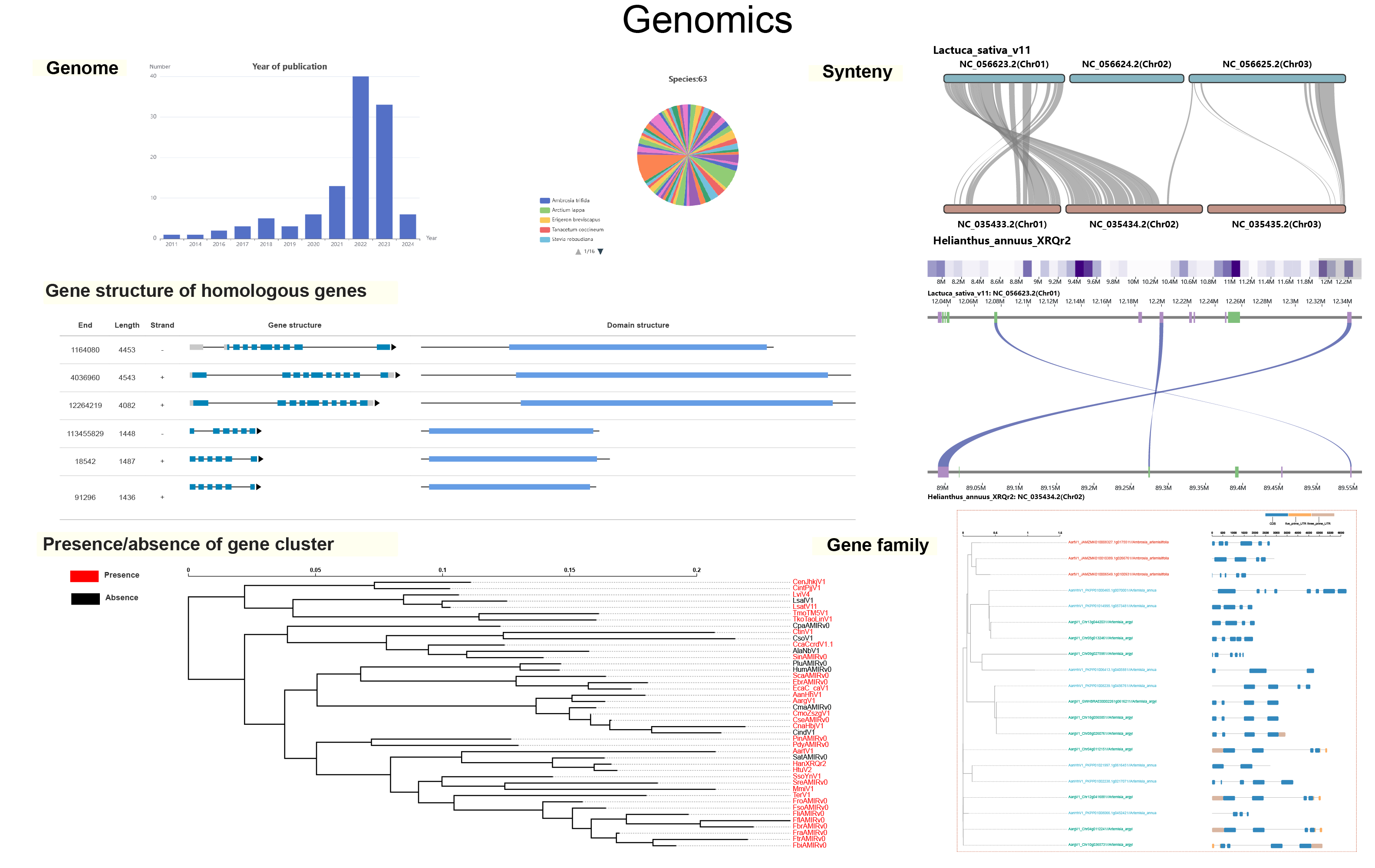
- Explore gene conservation(core, dispensable, and species-specific) and function using the cross-species super pan-genome.
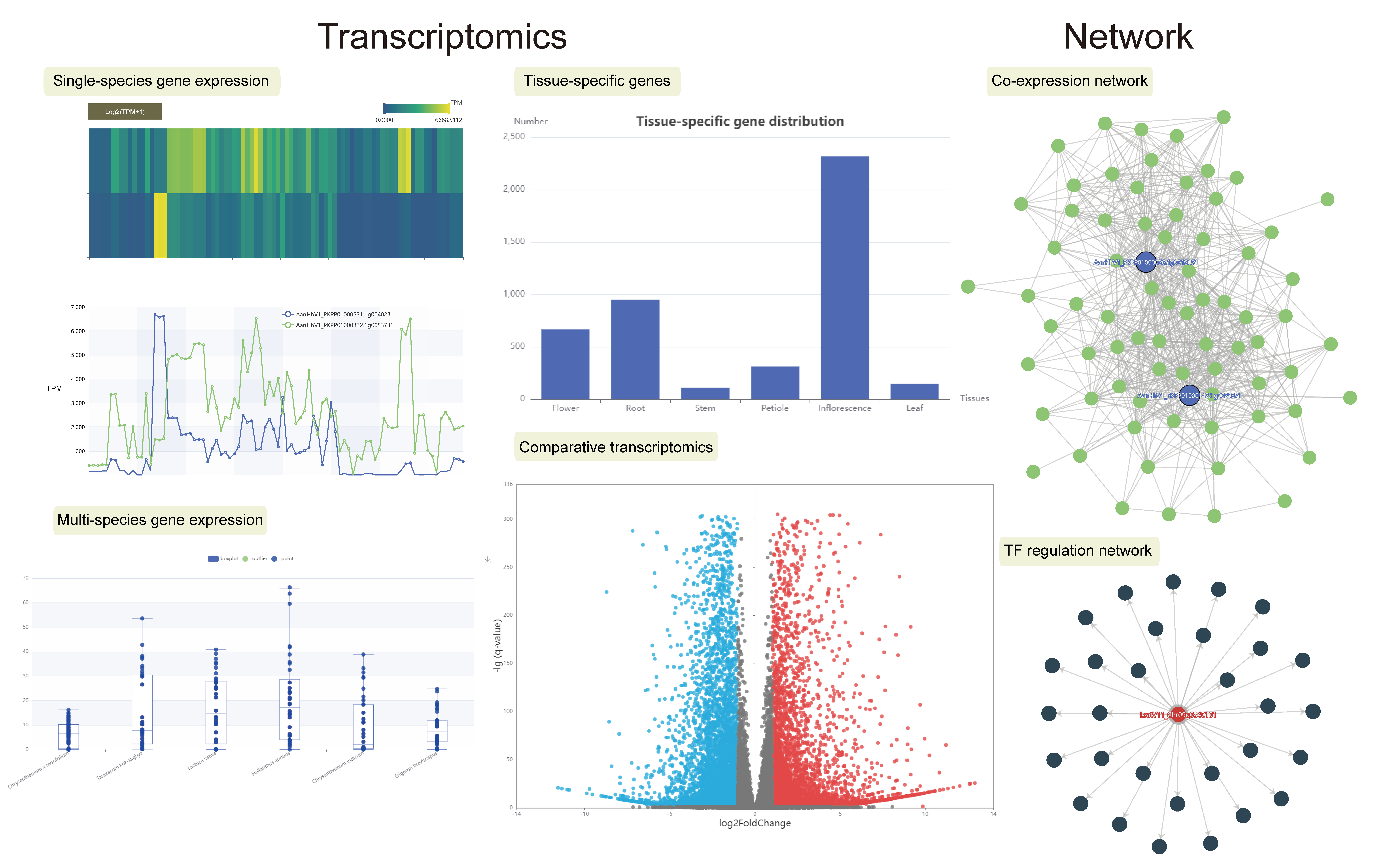
- Analyze and compare 3,897 gene expression profiles, identify tissue-specific genes, and mine differentially expressed genes related to key traits.
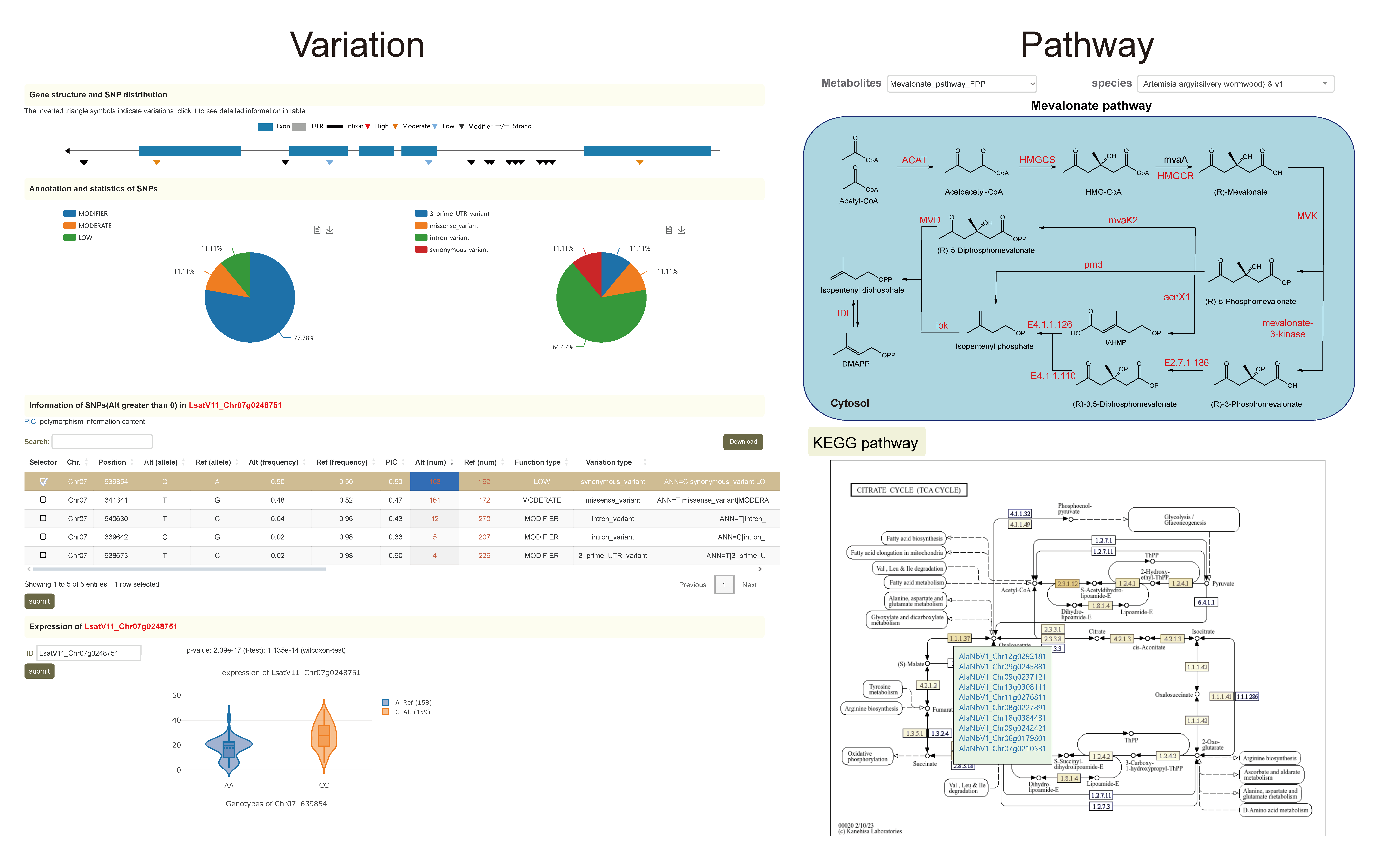
- Retrieve SNPs/InDels on important genes and mine variations and genes associated with important traits.
- Retrieve co-expressed genes and construct a gene regulatory network.
- Identify candidate genes involved in the biosynthesis of terpenoid, costunolide, artemisinin, chicoric acid and cyanidin.
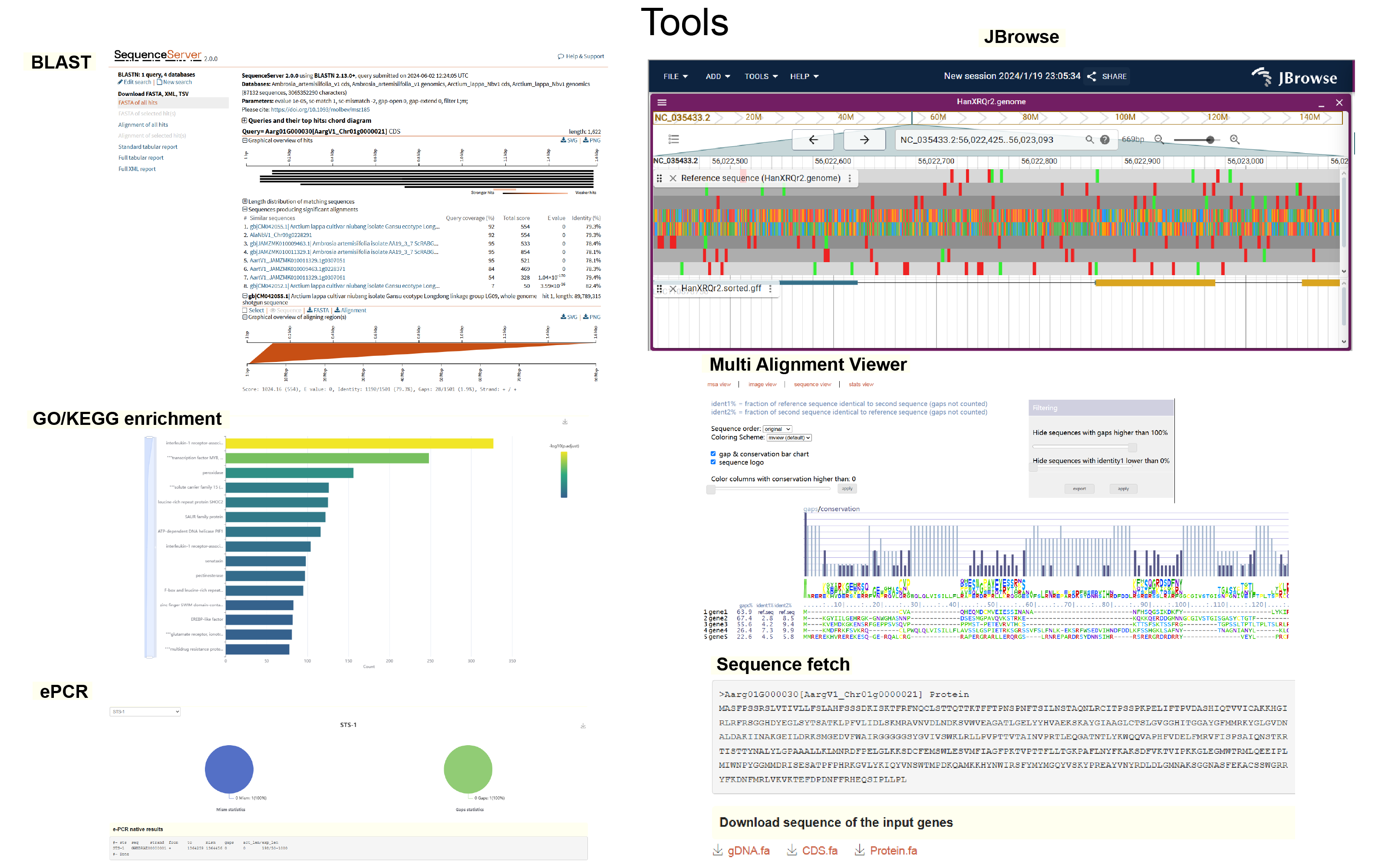
- Perform extensive customized bioinformatics analyses using tools module.
- Design primers for genes or positions of any species using the ‘Primer-design’ tool and assess primer specificity with ‘e-PCR’.
This assembly with annotation files
This assembly with predicted gene models
This assembly with RNA-seq data
Assembly level
- All the species are listed according to their positions on the species tree (see Download).
Overview
Protein Coding Gene Number
Genome Size (MB)
Our Resources
Portals of omics data & tools
Statistics of the number of genes |
|
How to cite us
Dongxu Liu, Chengfang Luo, Rui Dai, Xiaoyan Huang, Xiang Chen, Lin He, Hongxia Mao, Jiawei Li, Linna Zhang, Qing-Yong Yang, Zhinan Mei, AMIR: a multi-omics data platform for Asteraceae plants genetics and breeding research, Nucleic Acids Research;, gkae833, https://doi.org/10.1093/nar/gkae833